From correlative dead ends to functional discoveries—CRISPR screening gives research teams the power to ask questions that actually lead to actionable drug targets.
Introduction: Stop Guessing. Start Asking the Right Questions.
Your transcriptomics data lights up with differential expression. Dozens of genes—maybe hundreds—show promise on heatmaps. But when you validate them in vitro, nothing moves. The drug doesn’t work. The cells don’t die. The project stalls.
You’re not alone. This disconnect between gene expression and functional impact is a widespread pain point in drug discovery. Many labs have poured resources into genes that looked statistically significant but turned out to be biologically irrelevant.
That’s where CRISPR screening changes the game. Instead of chasing correlation, CRISPR screens allow you to ask: “What really matters for this phenotype?” Whether it’s cell survival, drug resistance, or immune evasion, functional genomics lets you find out—fast.
What Research Questions Can a CRISPR Screening Library Help You Answer?
Too often, teams begin with vague questions like “Which genes are differentially expressed?” But CRISPR screening pushes a more powerful agenda: “Which genes functionally drive the phenotype I care about?”
Here are the real-world research questions you can finally answer with a well-designed CRISPR screen:
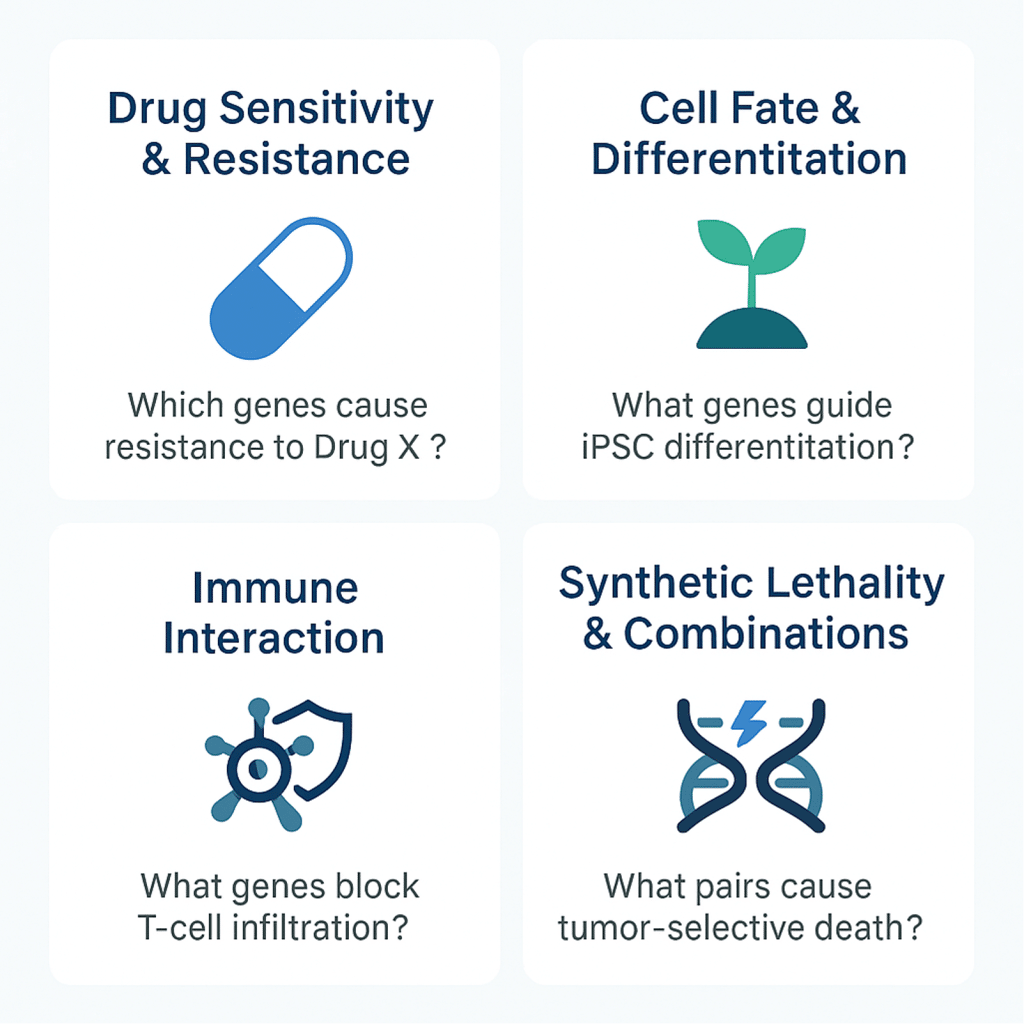
Drug Sensitivity & Resistance
• Which genes, when knocked out, sensitize cancer cells to a specific inhibitor?
• Which genes confer resistance when overexpressed (CRISPRa) or silenced (CRISPRi)?
Cell Fate & Differentiation
• What genes regulate pluripotency loss during iPSC differentiation?
• Which factors are essential for lineage commitment in organoid development?
Immune Interaction
• Which tumor genes block T-cell infiltration or activation?
• What host genes allow viral replication or immune evasion?
Synthetic Lethality & Combination Therapy
• What genetic pairs cause cell death only when inhibited together?
• Can I uncover dependencies unique to KRAS mutant tumors or p53-null backgrounds?
Each of these questions leads to real therapeutic strategies. CRISPR screens aren’t just hypothesis generators—they’re decision accelerators.
To explore available genome-wide and pathway-specific CRISPR screening libraries designed to address questions like these, click here.
From Exploratory Data to Actionable Discovery
Let’s illustrate the power of shifting from correlation to function.
A European oncology startup had invested months in RNA-seq, targeting the most upregulated genes in treatment-resistant cells. Yet, none of these targets reproduced their expected effects in knockout models. Frustrated and running out of runway, they pivoted.
They performed a pooled CRISPR knockout screen using Ubigene’s Human CRISPR KO Library v3.0. Surprisingly, the key resistance gene wasn’t overexpressed at all. It was a stress chaperone with tight regulation—missed entirely by transcriptomics but essential for drug response.
The screen saved the program. A new compound targeting the stress pathway is now advancing through preclinical validation.
This is the power of function-first screening. It tells you what actually matters—not just what’s loud in your data.
Functional Questions CRISPR Helps You Answer
| Functional Question | RNA-Seq | CRISPR Screen |
| Which genes cause resistance to Drug X? | ||
| What genes are essential for T-cell killing? | ||
| Can we find context-specific lethality in p53-null tumors? | ||
| What genes drive metastasis in a 3D model? |
To make CRISPR screening work for your study, phenotype selection is key. Ask yourself: What outcome can I reliably measure in my system?
Real Studies, Real Discoveries
- Immunotherapy Resistance Pathways
- Study: UCSF used genome-wide knockout libraries in tumor models.
- Finding: PTPN2 knockout significantly increased T-cell infiltration.
- Impact: Basis for new combination immunotherapies.
- Synthetic Lethality in Breast Cancer
- Study: Functional screen on BRCA1-mutant cells.
- Finding: Validated lethal interaction with PARP inhibition.
- Impact: Supported the development of Olaparib and related drugs.
- Host-Pathogen Interactions
- Study: SARS-CoV-2 screens identified host dependency genes.
- Finding: Lipid transport genes were critical to viral replication.
- Impact: Proposed druggable targets complementing vaccine efforts.
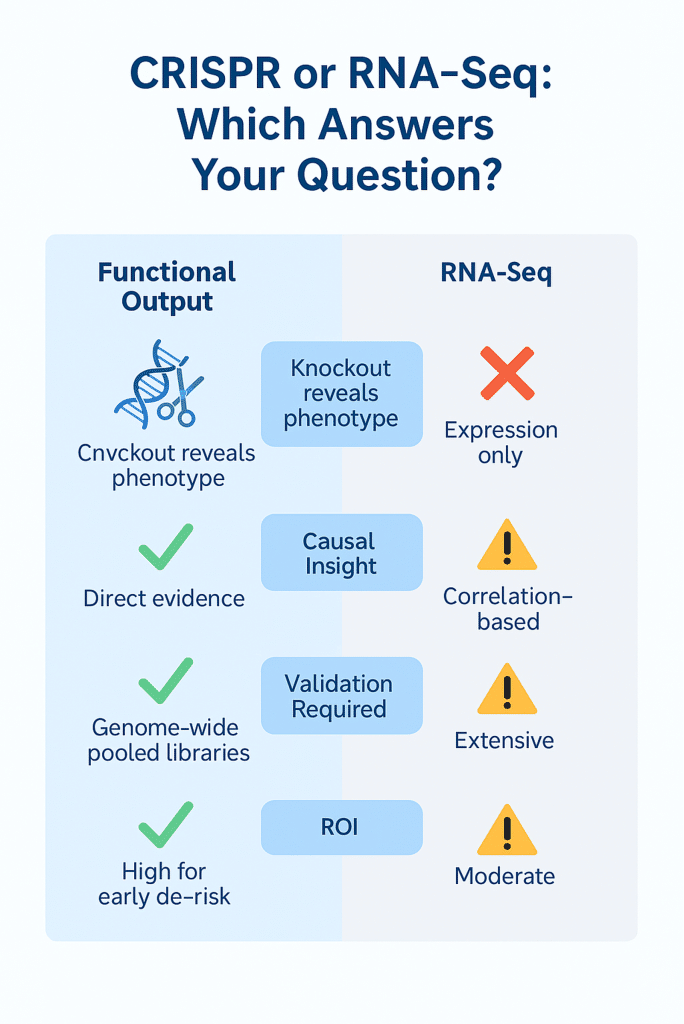
CRISPR Screening vs RNA-Seq: Why Function Beats Correlation
| Feature | CRISPR Screening | RNA-Seq |
| Functional Output | ||
| Causal Insights | ||
| Validation Requirement | ||
| Scalability | ||
| ROI for Drug Discovery |
RNA-seq still plays a role, but CRISPR screening delivers decisive data on gene function.
Common Mistakes (and How to Avoid Them)
| Mistake | Consequence | Fix |
| Using low MOI | Mixed editing, lost signal | Use MOI <0.3 with high-titer virus |
| No biological replicates | Poor reproducibility | Minimum 3 replicates per condition |
| Vague phenotype | Non-informative readout | Use measurable outcomes (viability, reporter) |
| No hit validation plan | Delayed timelines | Build in rescue/secondary assays |
Avoiding these pitfalls protects both your results and your budget.
Explore Ubigene’s CRISPR Gene Editing Tools for Functional Screening
Ubigene offers end-to-end CRISPR screening solutions optimized for drug discovery and functional genomics:
Human CRISPR KO Library v3.0
Covers 19,000+ genes with low off-target rates—ideal for broad essentiality profiling.
Oncology-Focused CRISPRi Library
Designed for suppression of essential genes in sensitive pathways.
Pre-validated Knockout Cell Lines
A549, HeLa, HCT116 and more—jumpstart your screen with robust, ready-to-use models.
For full access to Ubigene’s complete suite of CRISPR gene editing technologies, click here.
Designing the Right Phenotype: Where Most Screens Succeed or Fail
A powerful CRISPR screen begins not with gRNAs—but with a well-defined, measurable phenotype. Yet this is where many promising studies falter. If your screen’s readout doesn’t truly reflect the biological process you’re targeting, no amount of sequencing will rescue your results.
Ask yourself:
• Are you measuring something that directly links to your hypothesis (e.g., cell survival, reporter activation, fluorescence intensity)?
• Is the phenotype quantifiable and reproducible across replicates?
• Can it be detected using available methods—such as FACS, antibiotic selection, or microscopy?
Examples of robust phenotype designs:
• Positive selection: Surviving cells after drug treatment indicate resistance-related gene knockouts.
• Negative selection: Loss of fluorescence in a reporter line identifies regulators of a specific promoter.
• FACS enrichment: Sorting cells based on surface markers or fluorescent reporters allows for fine-grained screens in immune cell subsets.
Real case insight: A translational genomics team screening for genes that regulate PD-L1 expression designed a reporter cell line where PD-L1 was fused to GFP. This allowed real-time FACS sorting and enabled highly specific identification of regulators—without relying on antibodies or secondary assays.
Tip: If you can’t measure it clearly, you can’t screen it effectively. Work backwards from your disease model to define a phenotype that captures what matters most.
By refining phenotype selection, researchers not only improve hit quality—they also save time in downstream validation and interpretation.
Final Thought: The Best Question Wins
Drug discovery isn’t just about data—it’s about asking the question that saves time, money, and lives.
CRISPR screening empowers research teams to stop guessing and start validating. Instead of chasing noisy data, you can find out which genes actually matter and design better drugs—faster.
If you’re still relying on correlative tools alone, ask yourself:
What questions are you still not answering?
Maybe it’s time you did.